New Seminars Are Coming Soon
Past Seminars
Panel Discussion on Single Cell Multi-Omics: From Technology to Analysis
Explore the rapidly evolving world of single-nucleus omics—from transcriptomics and chromatin accessibility to multi-omic integration and downstream analysis. This panel brings together UCI experts to share insights on experimental platforms, cutting-edge computational tools, and collaborative strategies for accelerating discovery. Join us for brief presentations (three 15 minutes presentations), interactive discussions, and networking opportunities designed to foster cross-lab collaboration and drive innovation in single-cell biology.
The Genomics Research and Technology Hub


Modeling glioblastoma using genetically engineered brain organoids and spatial transcriptomics
10x Genomics and STEMCELL Technologies are excited to invite you to a webinar on the application of organoids and spatial transcriptomics in the study of Glioblastoma and development of a pre-clinical drug discovery platform.
Glioblastoma (GBM) is an aggressive, incurable form of brain cancer characterized by significant cellular heterogeneity due to interactions between tumor cell-intrinsic progenitor states and cell-extrinsic cues that promote rapid evolution. However, very few new therapeutic options for GBM have been developed in the last decade, highlighting the need for improved preclinical models. To address this need, we generated engineered GBM organoids (eGBOs) from genetically modified stem cells harboring mutations of both the Proneural and Mesenchymal subtypes of GBM. Using single-cell RNA sequencing (scRNAseq) and spatial transcriptomic analyses, we observed that GBM-associated mutations disrupt neurodevelopmental gene regulatory networks resulting in altered brain organoid morphology in vitro. Additionally, we found that transplantation of cells from eGBOs into mice results in tumors that recapitulate subtype-specific characteristics of human GBM tumors. Finally, we identified dynamic gene expression changes in developmental cell states underlying tumor progression using integrated single-cell transcriptomic data from eGBOs and patient GBM tumors. These single-cell and spatial transcriptomic analyses demonstrate that eGBOs can recapitulate GBM tumorigenesis and cellular heterogeneity found in patients. Further, this work provides an important validation of engineered cancer organoids as a human- relevant model that can enable future pre-clinical drug discovery of therapeutics designed to target patient-specific genetic backgrounds.

Experimental Design Sessions with 10x-perts
Please email christine.kao@10xgenomics.com to schedule your one-on-one appointment
Sign up for your individual session to discuss how the latest advancement in Chromium Single Cell,
Visium Spatial, and Xenium In Situ platforms from 10x Genomics can help you push the boundaries of
your research. Here are some updates from 10x, please email christine.kao@10xgenomics.com to
schedule an appointment.CRISPR Screening w/ GEM-X Flex
● Profile tens to thousands of sgRNAs simultaneously
● Directly link CRISPR edits and gene expression phenotypes at single cell resolution
● Per sample costs between $380 – $700 when multiplexing (min of 4 samples)
● CRISPR Screening with GEM-X Flex Gene Expression (technical note)
● Dissecting genetic networks with scalable and affordable single cell CRISPR screens (webinar)Visium HD 3′
● Species agnostic single cell scale spatial transcriptomics platform
● Compatible with Fresh Frozen and Fixed Frozen tissues
● >30 tissue types tested to date
● Limited transcript diffusion compared to competing products
● Pre-order available now!
● Visium HD 3′ PosterGEM-X Universal 3′ & 5′ – On Chip Multiplexing
● Load 4 individual samples per chip
● Capture 5k cells per barcoded sample
● Gel beads contain sample ID so that multiplexing can happen on the chip and during the
instrument run
● $560 per sample
● Link describing how the kit works
Illumina Lunch and Learn – The Latest Multiomic Innovations from Illumina
Join us for a seminar to learn how Illumina technology can help you uncover the answers to drive your research forward. During this event, Illumina experts will present an overview of next-generation sequencing (NGS), including recent application and platform updates, focusing on multiomics approaches and advancements in data analysis. We will focus on single-cell sequencing, the 5-base genome, proteomics, and Constellation Mapped Read technology.

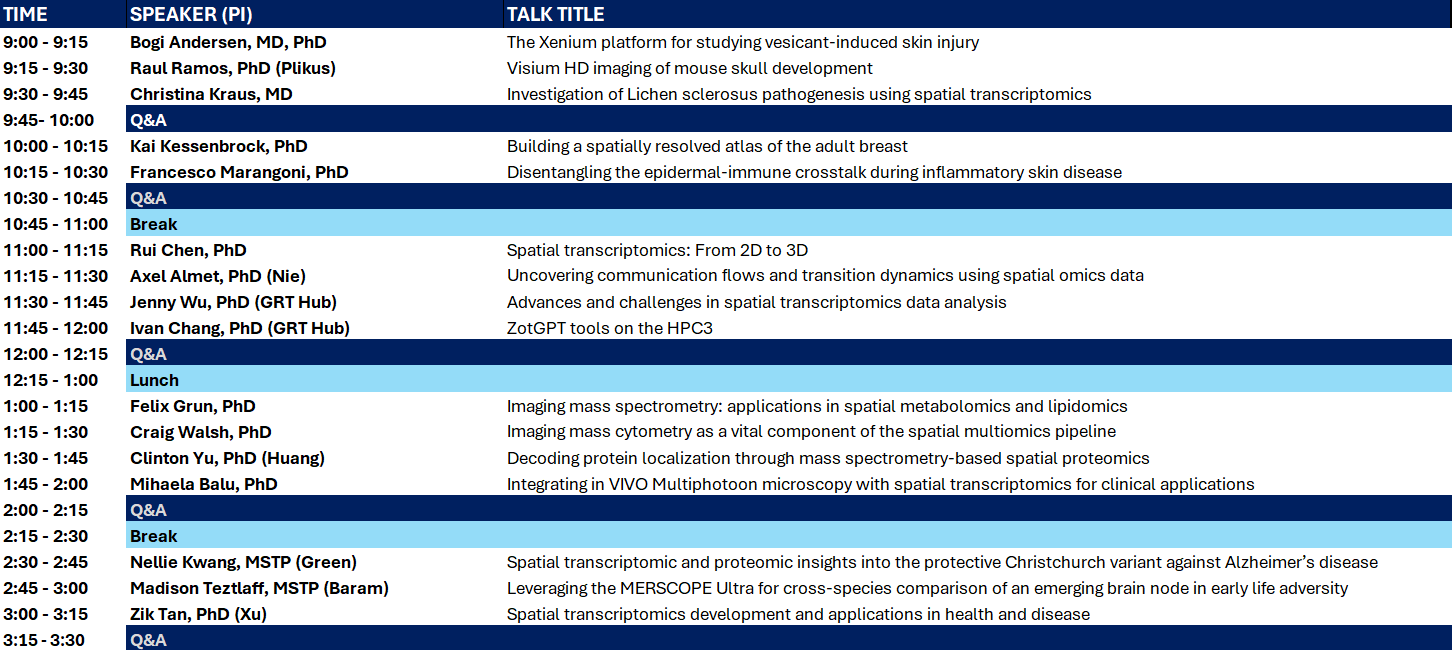
UCI Spatial Omics Symposium
We are excited to bring you “UCI Spatial Omics”, an in-person half day event to highlight recent developments in spatial omics research. Please join us for lively discussions of progress, challenges, and future applications.
The event is free, but registration is required. Please register by April 21, as space is limited.
The symposium is in partnership with…
- Genomics Research and Technology Hub
- Institute for Precision Health
- Chao Family Comprehensive Cancer Center
- Skin Biology Resource Center
- Center for Complex Systems Biology
QIAGEN Ingenuity Pathway Analysis Webinars
March 13: Interpretation of Spatial Transcriptomics Data using QIAGEN Ingenuity Pathway Analysis https://qiagen.zoom.us/webinar/register/3416674616219/WN_euTmFJnhQECyGvLJfGjVDg
March 17: IPA Deeper Dive: miRNA Investigation using QIAGEN Ingenuity Pathway Analysis https://qiagen.zoom.us/webinar/register/3416674616219/WN_yeU-YPsASgifSVtQWY-ScA
March 26: Network Construction and Customization – QIAGEN Ingenuity Pathway Analysis https://qiagen.zoom.us/webinar/register/3416674616219/WN_euizXh3HS8m-Mu9KU8kk3g
April 3: IPA New User Training https://qiagen.zoom.us/webinar/register/3416674616219/WN_Dl8L-NnTRiatSyT72MIYng
April 17: Biomarkers Investigation through Deep Curated ‘Omics Data Available through Ingenuity Pathway Analysis https://qiagen.zoom.us/webinar/register/2616964572539/WN_yQWjKwzJS32xGm7y1KcPgg
Genetics, Biomedical Computing and Genomics Seminar Series, “What’s New in Genomics Technologies?”
Presented by the Genomics Research and Technology Hub Staff
 PacBio Long – Read Technology Kinnex Bulk RNA and Single Cell Isoform Femto Pulse – Valentina Ciobanu, MSDownload
PacBio Long – Read Technology Kinnex Bulk RNA and Single Cell Isoform Femto Pulse – Valentina Ciobanu, MSDownloadUCI Special Seminar, “Huntington’s Disease Now – Mechanisms and Therapeutics”
Dr. Tabrizi is the Director of the University of College London Huntington’s Disease Centre and Joint Head of the Department of Neurodegenerative Disease at UCL Queen Square Institute of Neurology, a Principal Investigator at the UK Dementia Research Institute, and Honorary Consultant Neurologist at the National Hospital for Neurology and Neurosurgery.
She leads an internationally recognized basic bench science and translational research team focused on finding disease modifying therapies for Huntington’s Disease. She has received several awards, including the 2022 MRC Millennium Medal. In 2024, she was elected to the Fellowship of the Royal Society and to the US National Academy of Medicine.
“Advancements in Single-Cell Transcriptomics Through Long-Read scRNA-seq, Spatial Analysis, and GPT”, Rui Chen, PhD, UCI Department of Ophthalmology
Single-cell sequencing has revolutionized the molecular profiling of tissues and organs. However, current technologies still face several limitations and challenges. In this presentation, I will discuss various technologies we have developed to address some of these limitations. To explore mRNA alternative splicing at the single-cell level, we have employed single-cell RNA sequencing using both short-read and long-read high-throughput methodologies. Notably, we have identified a large percentage of novel isoforms, with many showing significant variation among cell types. Another limitation of scRNA-seq is the loss of spatial information. We have explored several single-cell spatial transcriptomics profiling technologies in the lab, and I will share our experiences with these methods. Finally, to address the need for more robust and effective tools for cell annotation in scRNA-seq data, we have utilized high-resolution single-cell reference datasets. By fine-tuning single-cell foundation models with domain-specific data, we have observed significantly improved performance compared to the initial models. These technological advancements can be readily adopted across various systems, further strengthening our single-cell toolboxes










